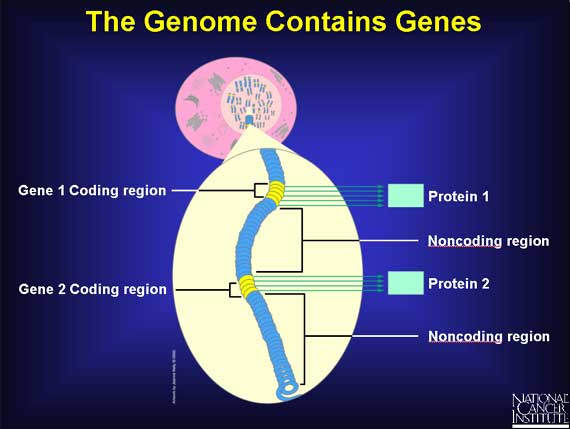
Genome Noncoding Regions: sequences that encode RNAs, introns, promoters and other control sequences, repeated sequences, and retroviruses. (Lewis, 207)
Non-coding "DNA sequences" (that) do not "code" for “amino acids.” Most non-coding DNA lies between "genes" on the "chromosome" and has no known function. Some noncoding DNA (sequences) play a role in the regulation of "gene expression." (NHGRI) (Includes) “introns” - sequences of DNA in the genes that are located between the “exons.” (MeSH) Scientist have recently discovered that the noncoding DNA in our genome, formerly considered “junk,” may play a major role in complex diseases by switching genes on and off. (Kandel4, 49) Editor's note 'noncoding' means does not code for the manufacture of a protein. Also referred to as 'noncoding DNA' 'noncoding RNA,' 'noncoding RNA genes,' 'non-protein encoding regions,' and 'junk DNA.’
Intron(s): part of a gene not used to manufacture proteins. (GNN) Non-coding sequence of DNA removed from "mature messenger RNA" prior to translation (GeneReviews) DNA sequence that interrupts the protein-coding sequence of a gene. (HGPIA) The sequence of DNA in between exons that is initially copied into RNA but is cut out of the final RNA transcript and therefore does not change the amino acid code. Some introns are known to affect gene expression. (NCI3) A portion of a gene that does not code for amino acids. The parts of the gene sequence that are expressed in the protein are called "exons," because they are expressed, while the parts of the gene sequence that are not expressed in the protein are called introns, because they come in between the exons. (NHGRI) A piece of the DNA in a gene which does not give raise to an "amino acid sequence." One or many blocks of sequence that are not represented in the mature RNA. During the process of ("RNA transcription") the base sequence of the gene forms a template for producing an mRNA molecule. Before the mRNA leaves the “nucleus” however, it is edited, and the base sequences forming introns are cut out. (Micklos, 83) They increase the probability of a “cross-over” event between genes. (Norman, 7/21/09) Also referred to as ‘intron sequence.’
Noncoding RNAs (ncRNAs): RNA which does not code for protein but has some “enzymatic,” structural or regulatory function. Excludes tRNA and rRNA. (MeSH) Also referred to as ‘untranslated RNA.’
Pseudogene: a sequence of DNA similar to a gene but nonfunctional; probably the remnant of a once-functional gene that accumulated mutations. (HGPIA) Resembles a gene but has been mutated into an inactive form over the course of “evolution.” A pseudogene shares an evolutionary history with a functional gene and can provide insight into their shared ancestry. (NHGRI) A copy of a gene that usually lacks introns and other essential DNA sequences necessary for function; pseudogenes, though genetically similar to the original functional gene, are not expressed and often contain numerous mutations. (GeneReviews) Similar in sequence to a particular protein encoding gene, and it may be transcribed into RNA, but it is not "translated" into protein. May be remnants of genes past, variants that diverged from the normal sequence too greatly to encode a working protein. (Lewis, 208)
Repeated Sequences: sequences of varying lengths that occur in multiple copies in the genome; represents much of the human genome. (HGPIA) Nucleotide sequences present in multiple copies in the genome. There are several types of repeated sequences. Tandem repeats are repeated copies which lie adjacent to each other. These can also be direct or inverted. The ribosomal RNA and transfer RNA genes belong to the class of middle repetitive DNA. (NCIt) Also referred to as ‘repeats’ ‘repeat sequences,’ and ‘repetitive DNA.’
Interspersed Repeat Sequences: sequences dispersed throughout the genome. (MeSH) Copies of "transposable elements" interspersed throughout the genome. (NCIt) Also referred to as 'interspersed repetitive sequences.
Microsatellite Repeats: a variety of simple repeat sequences that are distributed throughout the genome. They are characterized by a short repeat unit of 2-8 base pairs that is repeated up to 100 times. (MeSH) Repetitive DNA sequences usually several base pairs in length. Microsatellite sequences are composed of non-coding DNA and are not parts of genes. They are used as “genetic markers” to follow the "inheritance" of genes in families. (NHGRI) Multiple copies of the same base sequence on a chromosome; used as markers in physical mapping. (HGPIA) Repetitive segments of DNA scattered throughout the genome in non-coding regions between genes or within genes. Often used as markers for "linkage" analysis because of the naturally occurring high variability in repeat number between individuals. These regions are inherently unstable and susceptible to mutations. (GeneReviews) Also referred to as ‘microsatellites,' ‘microsatellite sequence,’ and 'satellite DNA.’
Transposable Elements: a class of DNA sequences that can move from one chromosomal site to another. (HGPIA) The most abundant type of repeat. Can move about the genome. Some encode “enzymes” that cut them out of one chromosomal site and integrate them into another. Comprise about 45% of the human genome sequence. Unstable transposons may lie behind inherited diseases that have several symptoms, because they insert into different genes. For example, “Rett syndrome.” (Lewis, 209) Discrete segments of DNA which can excise and reintegrate to another site in the genome. Most have not been found to exist outside the integrated state. (MeSH) You can think of transposons as 'selfish' genes, similar to viruses, which 'infect' a genome. In work that would earn her the Nobel Prize in 1983, American Barbara McClintock discovered transposable elements in 'maize,' where they altered the pigmentation of kernels. (Venter, 59) Also referred to as ‘transposons' and ‘jumping genes.’
Alu Sequence: a repeat about 300 bases long. (The) human genome may contain 300,00 to 500,000. Comprises 2 to 3 percent of the genome. We don't know exactly what these common repeats do. (Lewis, 208) The Alu sequence family is the most highly repeated interspersed repeat element in humans. “Transposition” of this element into coding and regulatory regions of genes is responsible for many heritable diseases. (MeSH) Also referred to as 'alu element.’
Tandem Repeat Sequences: copies which lie adjacent to each other, direct or inverted. (MeSH) A sequence of two or more DNA base pairs that is repeated in such a way that the repeats lie adjacent to each other on the chromosome. Tandem repeats are generally associated with noncoding DNA. In some instances, the number of times the DNA sequence is repeated is variable. Such variable tandem repeats are used in “DNA fingerprinting” procedures. (NHGRI) Also referred to as ‘tandem repeats.’
Minisatellite Repeats: tandem arrays of moderately repetitive, short (10-60 bases) DNA sequences which are found dispersed throughout the genome near “telomeres.” Their degree of repetition is two to several hundred at each "locus." Loci number in the thousands but each locus shows a distinctive repeat unit. (MeSH) Also referred to as ‘variable number of tandem repeats.’
Terminal Repeat Sequences: flank both ends of another sequence, for example, the long terminal repeats (LTRs) on retroviruses. (MeSH) Sequences that are repeated on both ends of a sequence, for example, the LTRs on "retroviruses." (NCIt) Also referred to as 'terminal repeats' and 'flanking repeats.'
Direct Repeat Sequences: repeats (occurring) in the same direction. (NCIt) Also referred to as ‘direct repeats.’
Inverted Repeat Sequences: copies of (DNA) sequences that are arranged in opposing orientation. (MeSH) Opposite to each other in direction. (NCIt)
Complementary Repeats: reads as the base complement in the opposite orientation. Complementary inverted repeats have the potential to form hairpin-loop or stem-loop structures when the complementary inverted repeats occur in double stranded regions. (MeSH)
Hyphenated Repeats: separated by some sequence that is not part of the repeat. (MeSH)
Palindromes: read the same backwards as forward. (MeSH)